Nucleotide sequence reagents are verifiable experimental reagents in biomedical publications, as a result of their sequence identities could be independently verified and in contrast with related textual content descriptors.
We have beforehand reported that incorrectly recognized nucleotide sequence reagents are attribute of extremely related human gene knockdown research, some of which have been retracted from the literature on account of attainable research fraud.
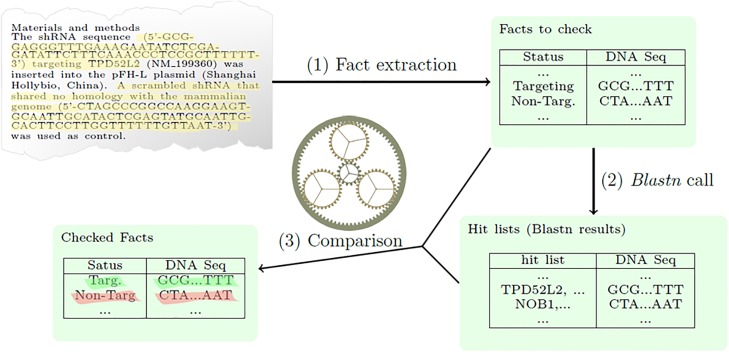
Because of the throughput limitations of guide verification of nucleotide sequences, we developed a semi-automated truth checking device, Seek & Blastn, to confirm the focusing on or non-targeting standing of revealed nucleotide sequence reagents.
From beforehand described and unknown corpora of 48 and 155 publications, respectively, Seek & Blastn accurately extracted 304/342 (88.9%) and 1066/1522 (70.0%) nucleotide sequences and a predicted focusing on/ non-targeting standing.
Seek & Blastn accurately predicted the focusing on/ non-targeting standing of 293/304 (96.4%) and 988/1066 (92.7%) of the accurately extracted nucleotide sequences. A complete of 38/39 (97.4%) or 31/79 (39.2%) Seek & Blastn predictions of incorrect nucleotide sequence reagent use have been right in the 2 literature corpora.
Combined Seek & Blastn and guide analyses recognized a listing of 91 misidentified nucleotide sequence reagents, which might be constructed upon by future research. In abstract, incorrect nucleotide sequence reagents characterize an under-recognized supply of error inside the biomedical literature, and truth checking instruments akin to Seek & Blastn could assist to establish papers and manuscripts affected by these errors.
Novel reagents for human prolactin research: large-scale preparation and characterization of prolactin receptor extracellular area, non-pegylated and pegylated prolactin and prolactin receptor antagonist.
To present new instruments for in vitro and in vivo prolactin (PRL) research, novel protocols for large-scale preparation of untagged human PRL (hPRL), a hPRL antagonist (del 1-9-G129R hPRL) that acts as a pure antagonist of hPRL in binding to hPRL receptor extracellular area (hPRLR-ECD), and hPRLR-ECD are demonstrated.
The interplay of del 1-9-G129R hPRL with hPRLR-ECD was demonstrated by aggressive non-radioactive binding assay utilizing biotinylated hPRL because the ligand and hPRLR-ECD because the receptor, by formation of secure 1:1 complexes with hPRLR-ECD beneath non-denaturing situations utilizing size-exclusion chromatography, and by floor plasmon resonance methodology.
In all three varieties of experiments, the interplay of del 1-9-G129R hPRL was equal to that of unmodified hPRL. Del 1-9-G129R hPRL inhibited the hPRL-induced proliferation of Baf/LP cells stably expressing hPRLR.
Overall, the organic properties of del 1-9-G129R hPRL ready by the protocol described herein have been much like these of the antagonist ready utilizing the protocol reported in the unique examine; nevertheless, the newly described protocol improved yields by>>6-fold.
To present long-lasting hPRL as a brand new reagent wanted for in vivo experiments, we ready its mono-pegylated analogue and located that pegylation lowers its organic exercise in a homologous in vitro assay.
As its future use would require the event of a PRL antagonist with extremely elevated affinity, del 1-9-G129R hPRL was expressed on the floor of yeast cells. It retained its binding capability for hPRLR-ECD, and this system was proven to be appropriate for future improvement of high-affinity hPRL antagonists utilizing a library of randomly mutated open studying body of del 1-9-G129R hPRL and deciding on high-affinity mutants by yeast floor show methodology.

