DNA polymerase is a ubiquitous enzyme that synthesizes complementary DNA strands in accordance with the template DNA in residing cells. Multiple enzymes have been recognized from every organism, and the shared capabilities of these enzymes have been investigated.
In addition to their elementary position in sustaining genome integrity throughout replication and restore, DNA polymerases are extensively used for DNA manipulation in vitro, together with DNA cloning, sequencing, labeling, mutagenesis, and different functions.
The elementary capability of DNA polymerases to synthesize a deoxyribonucleotide chain is conserved. However, the extra particular properties, together with processivity, constancy (synthesis accuracy), and substrate nucleotide selectivity, differ amongst the enzymes.
The distinctive properties of every DNA polymerase could result in the potential improvement of distinctive reagents, and due to this fact looking out for novel DNA polymerase has been one of the main focuses in this research area.
In addition, protein engineering strategies to create mutant or synthetic DNA polymerases have been efficiently creating highly effective DNA polymerases, appropriate for particular functions amongst the many varieties of DNA manipulations.
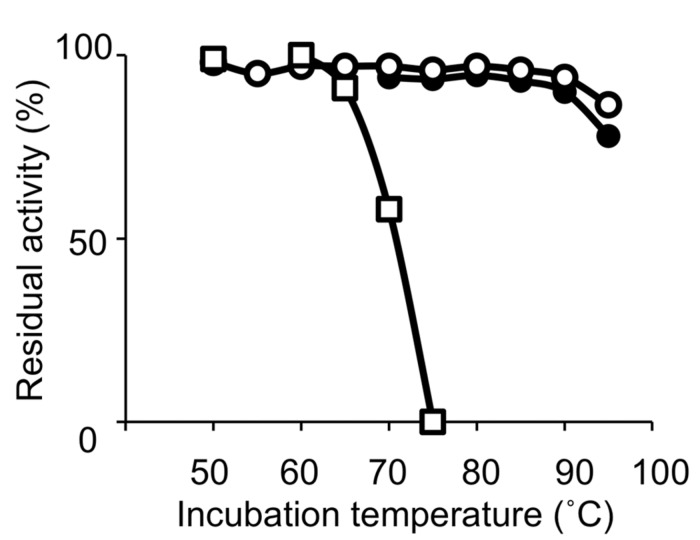
Thermostable DNA polymerases are particularly vital for PCR-associated strategies in molecular biology. In this assessment, we summarize the history of the research on creating thermostable DNA polymerases as reagents for genetic manipulation and talk about the future of this research area.
Development of proteome-broad binding reagents for research and diagnostics.
Alongside MS, antibodies and different particular protein-binding molecules have a particular place in proteomics as affinity reagents in a toolbox of functions for figuring out protein location, quantitative distribution and performance (affinity proteomics).
The realisation that the vary of research antibodies out there, whereas apparently huge is however nonetheless very incomplete and regularly of unsure high quality, has stimulated initiatives with an goal of elevating complete, proteome-broad units of protein binders. With progress in automation and throughput, a exceptional quantity of latest publications discuss with the sensible risk of deciding on binders to each protein encoded in the genome.
Here we assessment the necessities of a pipeline of manufacturing of protein binders for the human proteome, together with goal prioritisation, antigen design, ‘subsequent technology’ strategies, databases and the approaches taken by ongoing initiatives in Europe and the USA.
While the process of producing affinity reagents for all human proteins is advanced and demanding, the advantages of properly-characterised and high quality-managed pan-proteome binder sources for biomedical research, business and life sciences in normal can be monumental and justify the effort.
Given the technical, personnel and monetary sources wanted to fulfil this purpose, enlargement of present efforts could finest be addressed by giant-scale worldwide collaboration.